Re-analysis of client proteomics data from a third-party provider
We were asked by a client to re-analyse data generated from a proteomics study performed by another provider, to help them better understand and interpret the results.
The study design was as follows:
-
- Rodent plasma samples from two different treatment groups were depleted of albumin and IgG
- Samples were allocated across three multiplex TMT experiments
- Proteins were digested with trypsin and labelled with TMT isobaric labels, and combined into the three experiments.
- Fractions were generated using reversed-phase fractionation
- Each fraction was analysed by LC-MS/MS using an Orbitrap MS in DDA mode
- Data was analysed using a proprietary bioinformatics pipeline
The results previously obtained by our client were extensive, and included the selection of proteins of interest followed by principal component analysis to look for patterns of protein expression separating sample groups, and gene ontology (GO) analysis to investigate biological pathways of interest.
A volcano plot was used to show which proteins were of interest, selecting those that differed between the two treatment groups with a p-value below 0.01 and a log fold change beyond the median ± 2 MADs (median absolute deviations).
A list of the most interesting proteins (those with the highest and lowest fold changes) was provided.
The client asked for our support because neither the list of most interesting proteins or the GO terms provided seemed relevant to the biology being investigated, making it hard to interpret the results.
We took the peptide-level data our clients were provided with and started by analysing the reliability of the data. Firstly, there was some ion coalescence between some replicate trigger samples (high abundance controls used to increase detection of proteins of interest), but that had little impact on the data from the low abundance test samples. Next, we investigated the reproducibility of the sample data, although this was limited to investigation of a single duplicate due to the experimental design. While there was, unsurprisingly, an association between the abundance of precursors in the two samples, the performance is markedly lower than we see with our patent-protected DDA ProQuant® platform (comparison data is from human serum samples).
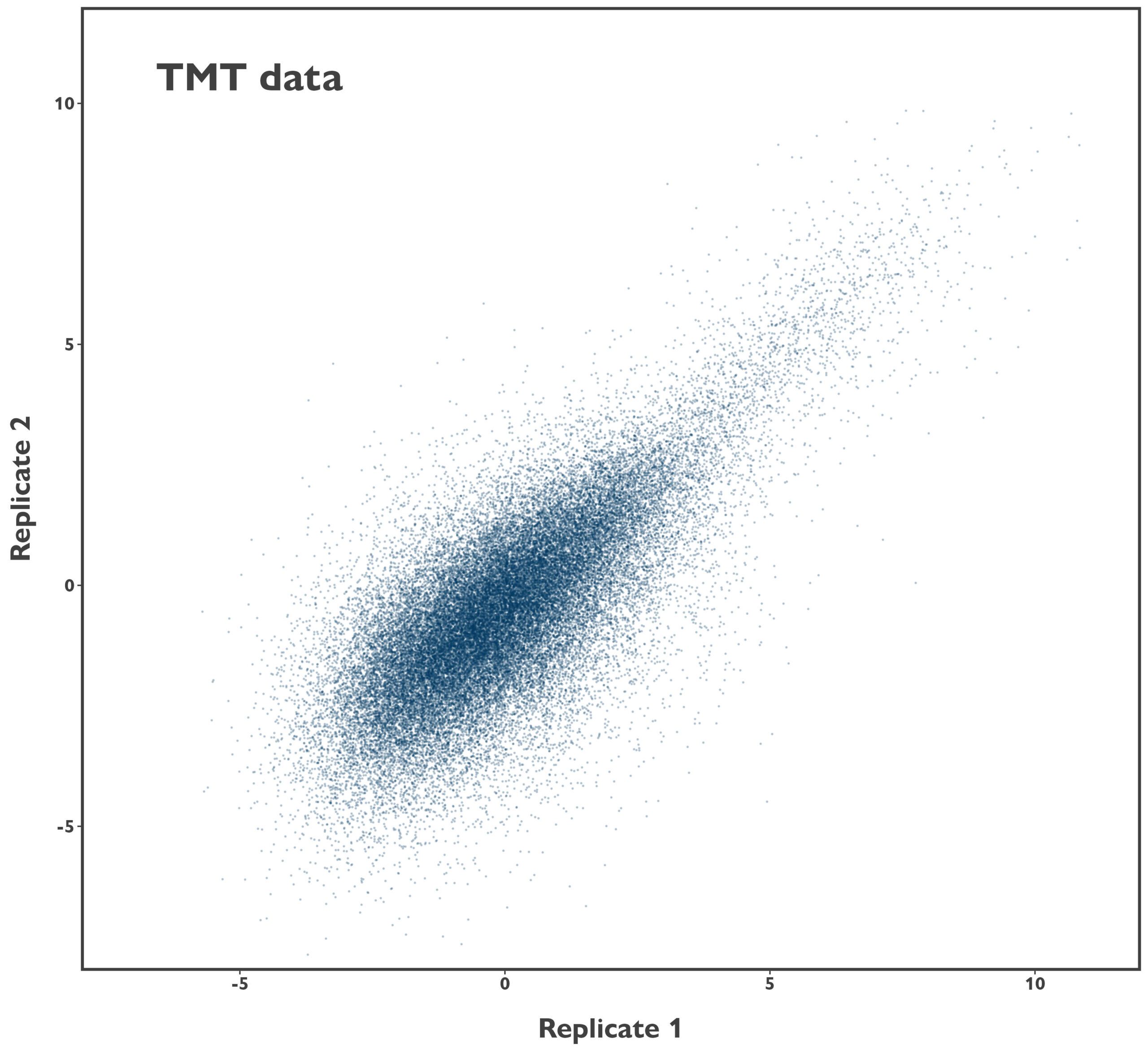
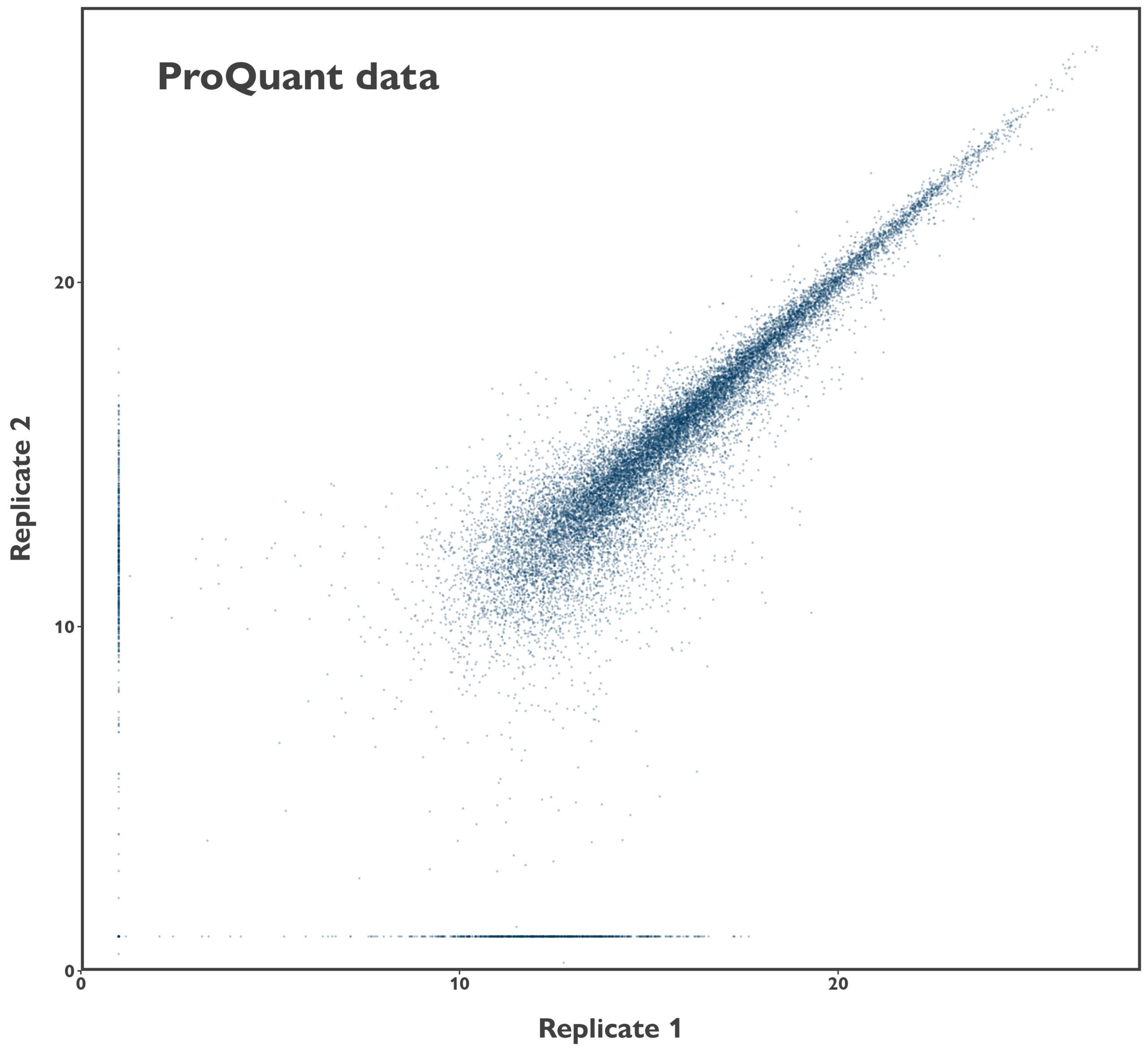
Figure 1: Comparison of sample reproducibility between competitor TMT data and ProQuant DDA data
Nevertheless, the data appeared to be suitable for further analysis.
The next stage of our review of the data was to revisit the statistical analysis. We took the same processed data generated by the original provider, but filtered for proteins of interest using our usual approach.
The original analysis restricted proteins to those whose association with treatment were significant at p < 0.01 (with as far as we can tell the cut-off chosen arbitrarily) and then again to those proteins with a minimum fold-change between the treated and untreated groups. By contrast, we first limited the analysis to only consider proteins which were quantified from at least two unique peptides, to improve the robustness of the results. The original analysis did not contain this key step, and over 80% of the proteins highlighted in the results as proteins of interest were identified from only a single peptide. Next, we restricted proteins to those remaining significant at an FDR of 10%, and did not limit to only those with a minimum fold-change between groups. Having carried out these alternative restrictions we listed the top ten most interesting proteins (most significant, not greatest fold change), and then carried out our own gene ontology analysis.
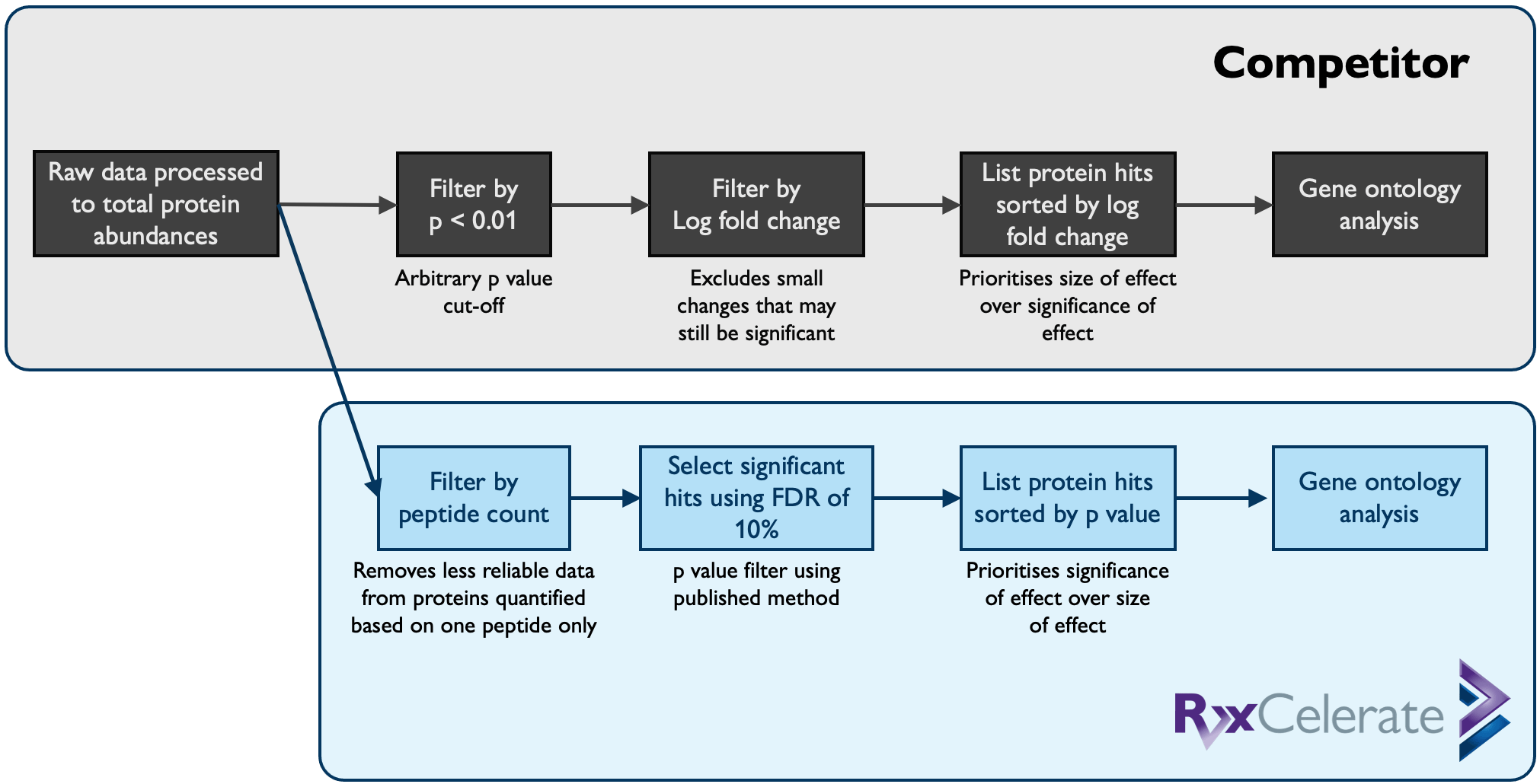
Figure 2: Data processing and statistical analysis pipelines
Having carried out the statistical analysis differently, we then presented the results to the client. There were immediately some key insights.
Firstly, the second protein on our list of hits was a relatively obscure protein whose UniProt function explicitly stated that it was involved in the signalling pathway of interest to the client.
Secondly, the gene ontology analysis was also interesting. While none of the biological processes were found to be significantly associated with treatment group (at an FDR of 10%), three of the top 20 most significant biological processes (all unadjusted p < 0.01) were directly related to the pathway targeted by the client’s test compound.
The client was very happy with the results we provided to them, and it gave them confidence that the test compound administered was affecting the biological pathway they intended it to.
This study highlights clearly that an enormous number of decisions get made throughout the generation of complex proteomics datasets, and getting experimental design and statistical analysis right is as important as the core technical proteomics analysis.
At RxCelerate we pride ourselves on making considered decisions throughout a study, helping our clients come to a balanced view of the results obtained from the work carried out for them.
Find out more about our Proteomics Services.
